Genomics: Insight

On Our Way to Understanding Autism Spectrum Disorders (ASD) Genetic Architecture

Autism Spectrum Disorders (ASD) are a group of neurodevelopmental disorders characterized by cognitive phenotypes such as restricted interests, repetitive behaviors and language and communication deficits. There is an enormous phenotypic heterogeneity observed in patients that is mirrored in a great number of genetic variants identified for ASD, with no single one explaining more than 1% of cases at best. In the past decade important advances have been made in identifying rare variants associated with ASD risk, and several genes and copy number variant (CNV) loci are now under scrutiny to elucidate the molecular mechanisms associated with ASD pathophysiology. However, despite the knowledge that common variation greatly contributes to ASD risk [1], the search for ASD-associated single common risk variants (SNPs) has not been as successful, largely because of limitations in the statistical power of previous ASD genome-wide association studies (GWAS).
A recent study in Nature Genetics finally sheds some light on the issue [2]. By greatly increasing the sample size compared to previous analysis, the authors were able to identify single common risk variants for ASD. Moreover, the study demonstrated that ASD shares a substantial amount of risk variants with other psychiatric disorders such as schizophrenia (SCZ) and depression, something that was long time hypothesized due to the high comorbidity observed between these disorders.
"A shared etiology between ASD and other psychiatric disorders such as schizophrenia (SCZ) and depression was demonstrated."
The study was based on a Danish population cohort with more than 30000 individuals, in which a third of those were ASD cases [3]. Full clinical records were available for the cohort, which allowed for separation in subgroups with different cognitive phenotypes (ASD with or without Intellectual Disability (ID), Asperger Syndrome diagnosis or other comorbidities, etc.). The analysis of this cohort initially identified significant genome-wide markers in three different loci. Then, replication data was obtained using five additional cohorts from European ancestry, and the expansion of the dataset with this additional batch of patients allowed for the identification of new common ASD risk variants in two additional loci.
After this initial discovery, the authors assessed the degree of genetic correlation between ASD and other cognitive traits such as SCZ, depression and educational attainment. A strong correlation was found, with varying degrees between different ASD categories, and in comparison no correlation was found with other traits such as height or mass body index, with no expected links to ASD. This findings allowed to explore those GWAS studies to discover new loci shared between ASD and this cognitive traits, concluding in the identification of an additional seven loci. In total, a total of twelve genome-wide significant loci are now confidently associated with ASD.
The identification of these group of confident SNPs, allowed for the calculation of the ASD heritability within different phenotypic categories (i.e., ASD patients with or without ID). Heritability is a concept used in population genetics that corresponds to the fraction of a given trait that can be explained by genetics. So, for example, a monogenic disease like Tuberous Sclerosis would have close to 100% heritability (if you have the specific mutation, you are almost guaranteed to have the disease), but traits like height are way more influenced by environment and therefore its heritability is of lower value. Thus, it was estimated that ASD without ID had a higher heritability than with ID. Interestingly, within different subgroups of ASD, Asperger syndrome appeared to have close to double heritability than the others.
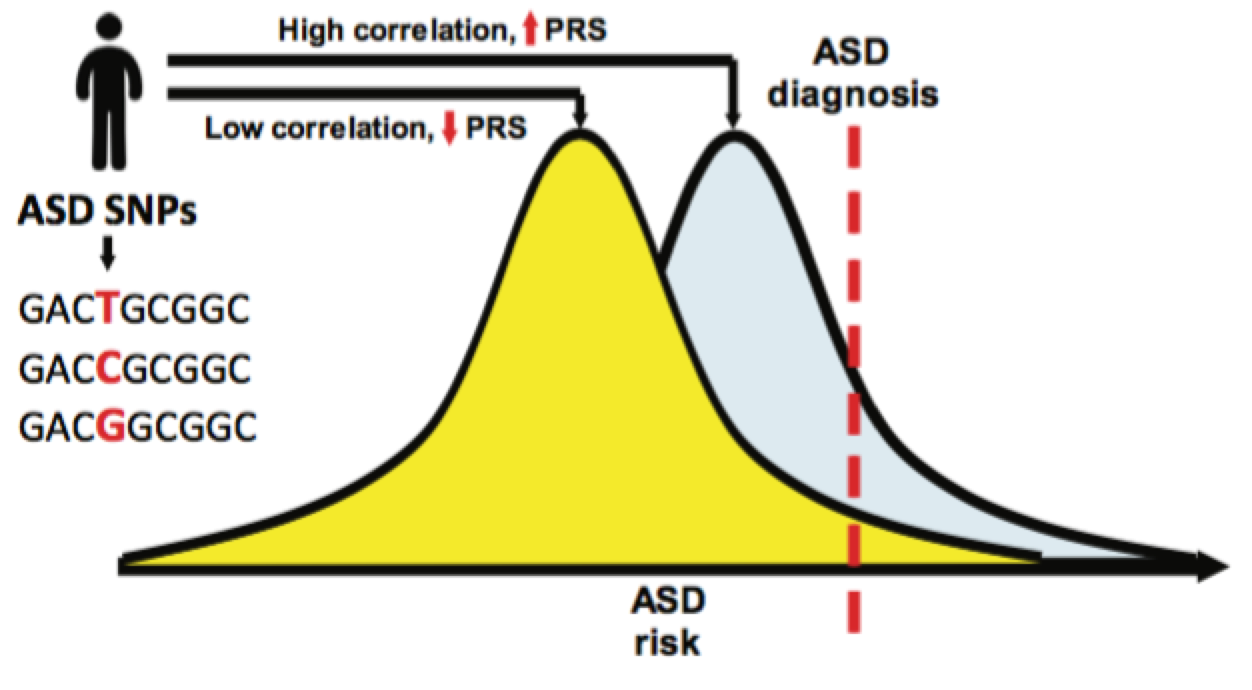
Most importantly, the identification of ASD SNPs also allows for the calculation of its polygenic risk score (PRS). PRS is a tool of promise in the new filed of personalized medicine, and consists in the estimation of risk in a patient on the basis of the correlation between the given trait SNPs and the patient genetic variants. The comparison of PRS scores between ASD subgroups and other cognitive measurements revealed a big difference, demonstrating that the genetic architecture between ASD subgroups is indeed heterogeneous.
Functional enrichment of the identified loci showed that they are located in conserved DNA regions and enhancer sequences, which are regulatory elements that control gene expression. Additional analysis of these enhancer-associated marks revealed that they are enriched in the cerebral cortex and in neuronal cell lines, especially in the developing brain. Furthermore, the genes associated with the confident ASD SNPs were mostly associated with neuronal functions and transcription factors, and they were most expressed during fetal corticogenesis. These observations are especially important as they are in agreement with previous findings on rare variation in ASD. For example, the most common rare CNV mutation found in ASD patients, the 16p11.2 loci deletion/duplication, has been associated with dysregulation of multiple signaling pathways, specifically on layer 3-4 cortical neurons during early brain development [4].
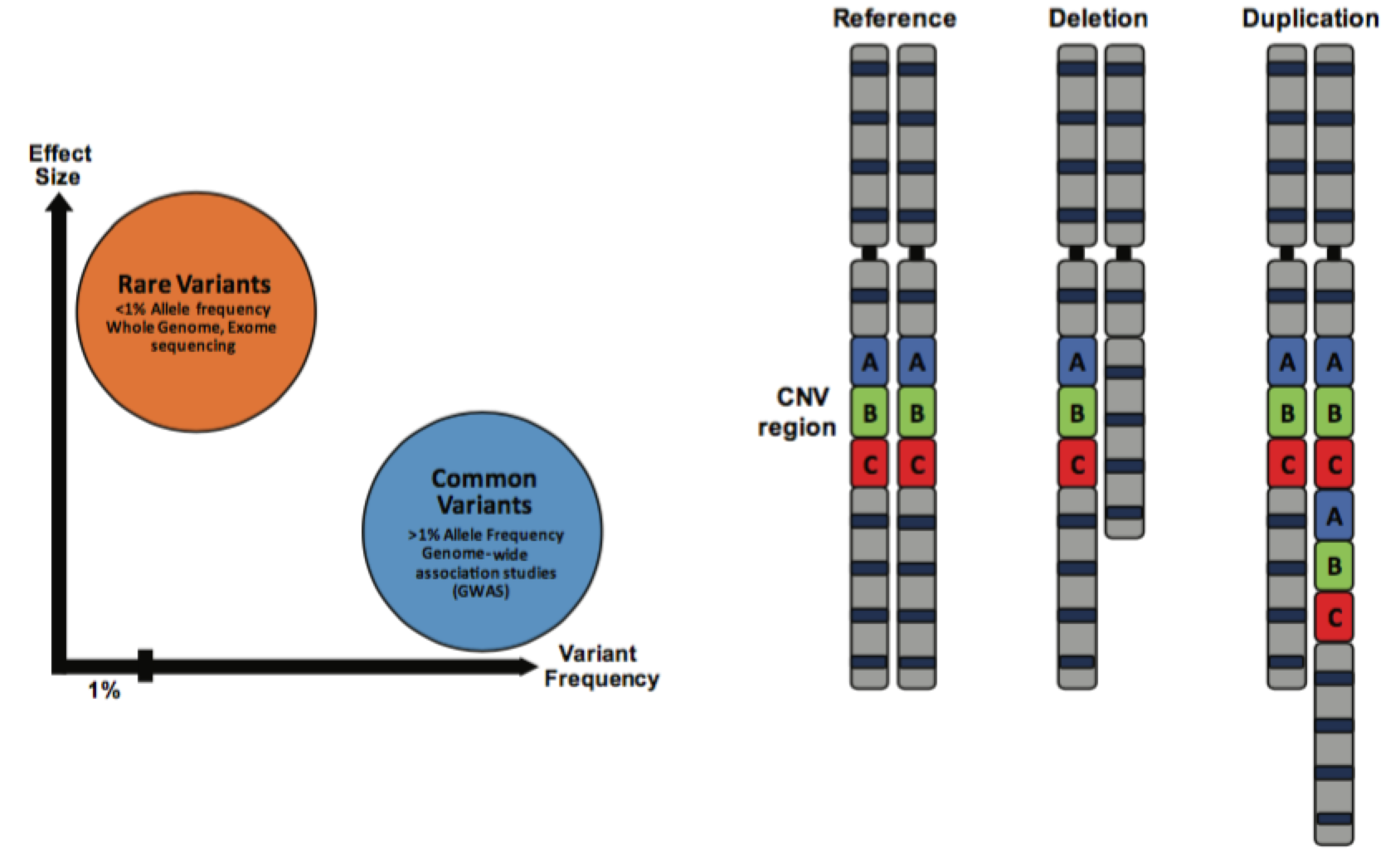
The most accepted hypothesis about ASD etiology to date is that ASD is a neurodevelopmental disorder that starts at early stages of brain development. Despite the large genetic heterogeneity observed, with hundreds of risk variants identified in dozens of different genes or CNVs, there is a convergence in the functional pathways that are most impacted [5]. A lot of the risk genes associated with ASD to date usually fall under one of the following categories; associated with neuronal functions, ranging from neuronal progenitor proliferation and differentiation to synaptic formation and maintenance, or associated with transcription and chromatin remodeling. One example of the former group would be SCN2A, a voltage-gated sodium channel responsible for the generation and propagation of action potential in neurons, and one example of the latter group would be CHD8, which regulates the transcription of several genes.
"Despite the large genetic heterogeneity observed (in ASD), […] there is a convergence in the functional pathways that are most impacted."
The observation that the first successful GWAS study on ASD identifies a subset of variants that can be associated with genes with similar properties as those previously identified, should be encouraging and exciting for scientists in the field working on characterizing the molecular pathways impacted. Moreover, the fact that we reached the threshold of sample size that allows for enough statistical power for variant identification paves the path for new analysis on different defined subgroups, such as whether people respond to a given treatment. This type of analysis has been successfully applied to other psychiatric disorders such as schizophrenia [6].
Taken together, the combination of advances on both genetic and molecular findings in ASD etiology will surely enhance the discovery and improvement of treatments and therapies. A better understanding on the genetic ASD causes along with current efforts on personalized medicine with genetic tests will help patients and clinicians to find the most appropriate treatment for each case. Combined with new findings in the molecular pathophysiology of ASD that facilitate drug discovery, we are bound to observe a great change in the ASD field in the following years.
References
[1]Gaugler, T., et al., Most genetic risk for autism resides with common variation. Nat Genet, 2014. 46(8): p. 881-5.
[2]Grove, J., et al., Identification of common genetic risk variants for autism spectrum disorder. Nat Genet, 2019. 51(3): p. 431-444.
[3]Pedersen, C.B., et al., The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry, 2018. 23(1): p. 6-14.
[4]Lin, G.N., et al., Spatiotemporal 16p11.2 Protein Network Implicates Cortical Late Mid-Fetal Brain Development and KCTD13-Cul3-RhoA Pathway in Psychiatric Diseases. Neuron, 2015. 85(4): p. 742-54.
[5]Satterstrom, F.K., et al., Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. bioRxiv, 2019: p. 484113.
[6]Yu, H., et al., Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry, 2018. 5(4): p. 327-338.
About the Author

Jorge Urresti is a postdoctoral researcher in the Department of Psychiatry in the University of California, San Diego. He received his PhD in Molecular Biology in the Autonomous University of Barcelona. His research focuses on elucidating the functional impact of ASD mutations using patient-derived cells, and developing stem-cell applications for modeling neurodevelopmental disorders.