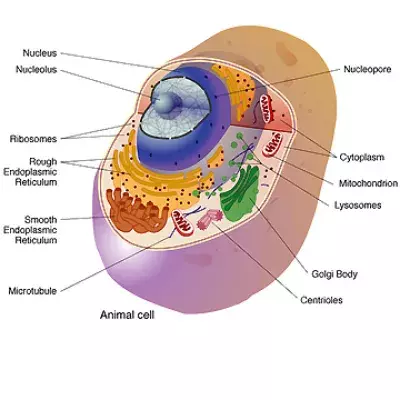
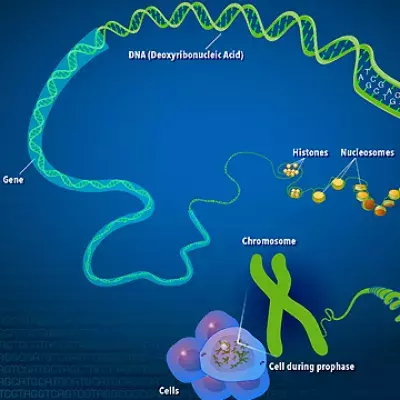
Resource Library
Error message
The page you requested does not exist. Please see resources related to "explore OR genome OR within OR us OR feature OR story OR sequencing OR big OR c".No Results were found. Please try refining your search or check out our Most Popular Resources below: